Lennard-Jones Potentials for the Interaction of CO2 with Five-Membered Aromatic Heterocycles | The Journal of Physical Chemistry A
![ANN] CellListMap.jl: short-ranged particle pairwise computations (lennard- jones, gravitational, etc) - Package announcements - Julia Programming Language ANN] CellListMap.jl: short-ranged particle pairwise computations (lennard- jones, gravitational, etc) - Package announcements - Julia Programming Language](https://global.discourse-cdn.com/business5/uploads/julialang/original/3X/b/f/bf80b823f430abde0bc8b36d60d1286ba461871a.png)
ANN] CellListMap.jl: short-ranged particle pairwise computations (lennard- jones, gravitational, etc) - Package announcements - Julia Programming Language

Lennard-Jones Potentials for the Interaction of CO2 with Five-Membered Aromatic Heterocycles | The Journal of Physical Chemistry A

Fast and Flexible GPU Accelerated Binding Free Energy Calculations within the AMBER Molecular Dynamics Package | bioRxiv

GPU-accelerated molecular dynamics: State-of-art software performance and porting from Nvidia CUDA to AMD HIP - Nikolay Kondratyuk, Vsevolod Nikolskiy, Daniil Pavlov, Vladimir Stegailov, 2021

PDF) Comparison between parallel and distributed molecular dynamics simulations of Lennard-Jones systems | Dorian Gorgan - Academia.edu



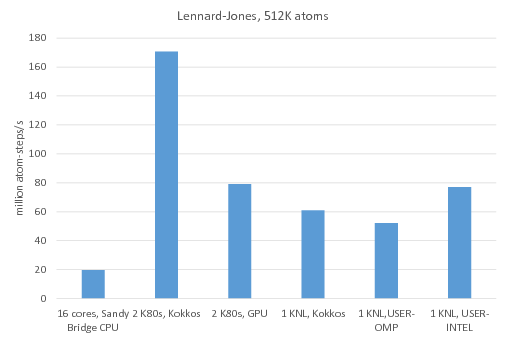
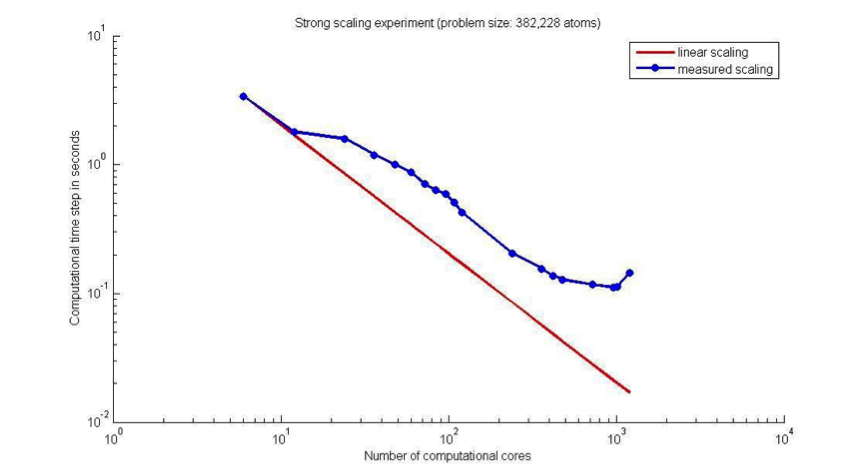
![exercises:2017_ethz_mmm:pythonmd [CP2K Open Source Molecular Dynamics ] exercises:2017_ethz_mmm:pythonmd [CP2K Open Source Molecular Dynamics ]](https://www.cp2k.org/_media/exercises:2017_ethz_mmm:screen_shot_2017-03-03_at_05.47.25.png?w=600&tok=47f5d6)

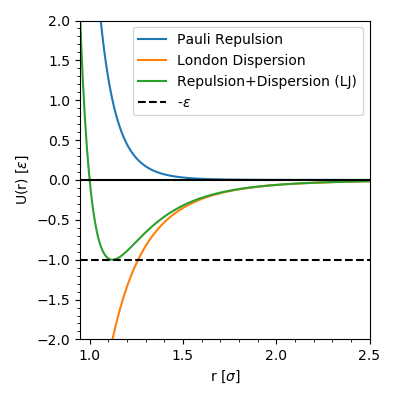

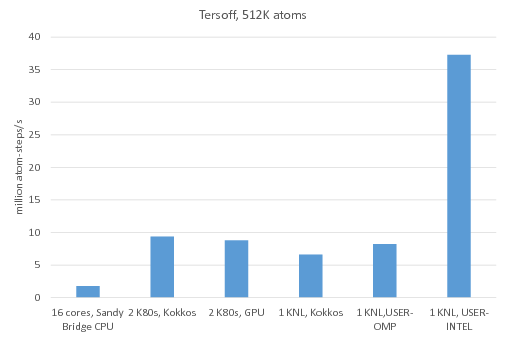
![PDF] GPU-accelerated Gibbs ensemble Monte Carlo simulations of Lennard-Jonesium | Semantic Scholar PDF] GPU-accelerated Gibbs ensemble Monte Carlo simulations of Lennard-Jonesium | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/76d4df3af9b558d2206a300e6803e9afabe28bb9/5-Figure4-1.png)